In mammals, female (XX) and male (XY) cells exhibit sex chromosome dimorphism. To achieve dosage compensation of X-linked genes, female cells undergo X chromosome inactivation (XCI) during early embryo development. Abnormal XCI leads to severe developmental defects such as intellectual disabilities in humans and even embryonic lethality. Hence, investigating the mechanisms of XCI is of both scientific and clinical significance. Inactive X chromosome (Xi) adopts a microscopically distinguished condensed conformation, known as the “Barr body”. Previous studies based on the mESC differentiation system found that TADs and compartments are significantly weakened, and two “megadomains” separated at the Dxz4 macrosatellites occurred on the Xi. Wei Xie’s lab and collaborators have previously conducted a series of studies on the dynamics and functions of three-dimensional chromatin architecture in animal reproduction and early development including spermatogenesis (Wang et al., Mol Cell, 2019), oogenesis (Du et al., Mol Cell, 2020), preimplantation embryos (Du et al., Nature, 2017), postimplantation embryos (Zhang et al., Nat Genet, 2018; Xiang et al., Nat Genet, 2020), and somatic cell nuclear transfer (SCNT) embryos (Zhang et al., Mol Cell, 2020). However, the dynamic changes, mechanisms, and functions of the chromatin architecture during XCI in female embryo development remain elusive.
On September 10th, a joint team led by Prof. Wei Xie and Assistant Prof. Haifeng Wang at Tsinghua University and Prof. Sundeep Kalantry at the University of Michigan Medical School published a research paper in Nature Genetics entitled Stepwise de novo establishment of inactive X chromosome architecture in early development. This study identified a unique Xist-separated megadomain structure (X-megadomains) on the Xi and revealed the transcriptional regulation to achieve both essential gene activation and global silencing during the early stage of X chromosome inactivation.
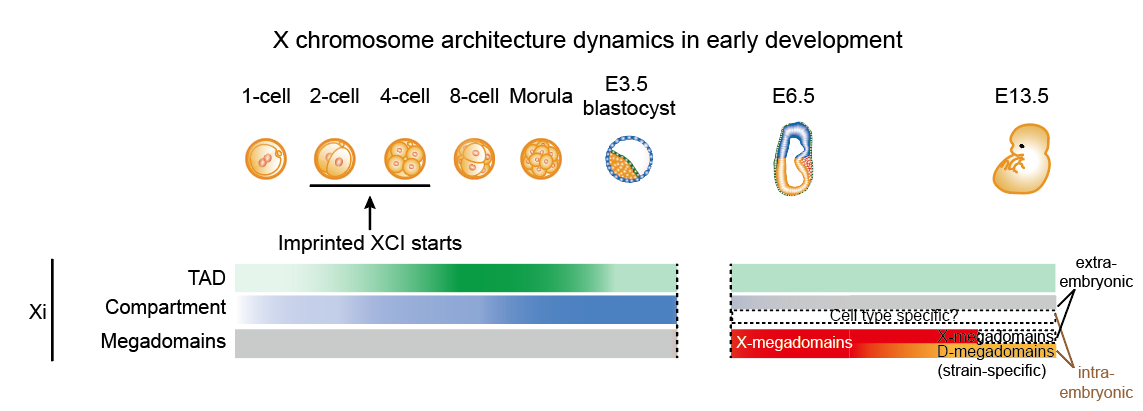
Fig 1. X chromosome architecture dynamics in early development
In mice, the paternally inherited X chromosome (Xp) is selectively silenced. Such imprinted XCI initiates around the four-cell stage and the inactivation of the Xp is stably maintained in extraembryonic tissues. By contrast, Xp in embryonic epiblast cells is reactivated, followed by random inactivation of one of the two X chromosomes (random XCI). This study systematically examined the 3D chromosome structure of the X chromosome across the early mouse development from 1-cell embryos to E9.5 extraembryonic cells and E13.5 embryonic cells, spanning the establishment and maintenance of imprinted XCI and random XCI. The researchers revealed that while TADs and compartments diminished from the Xi, a unique Xist-separated megadomain structure (X-megadomains) emerged on the Xi in mouse extraembryonic lineages, and transiently in the embryonic lineages. Previously reported Dxz4-delineated megadomains only appeared at later stages in some mouse strains but were completely absent in other mouse strains. X-megadomains were also observed in in vitro cultured extraembryonic endoderm (XEN) cells and were validated by 3D DNA/RNA FISH.
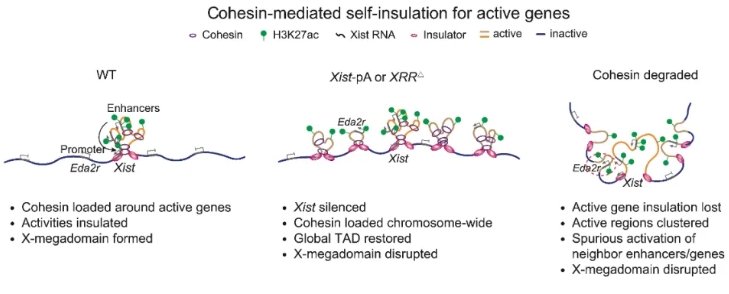
Fig 2. Cohesin-mediated self-insulation for active genes on the Xi
The researchers further revealed that X-megadomain boundary coincided with strong enhancer activities and cohesin binding in an Xist regulatory region (XRR). Xist regulatory region disruption or cohesin degradation impaired X-megadomains in extraembryonic endoderm cells and caused ectopic activation of regulatory elements and genes near Xist. The XRR region is enriched for H3K27ac, NIPBL (the Cohesin loader), and cohesin. Given that acetylated histones can recruit their reader BRD4 which interacts with NIPBL, the researchers propose a model that regulatory elements near active genes, for example, Xist, facilitate cohesin loading, which in turn insulates the regulatory activities within the local territories and promotes X-megadomain formation, thus achieving both essential gene expression and preventing aberrant activation of nearby genes that should be silenced. Taken together, this study unveils the dynamic reorganization of chromosome architecture during the de novo establishment of XCI in early mouse development and further sheds light on the potential function of the 3D genome in balancing gene silencing and selective activation of essential genes.
Prof. Wei Xie and Assistant Prof. Haifeng Wang from the School of Life Sciences of Tsinghua University and Prof. Sundeep Kalantry from the University of Michigan Medical School are the corresponding authors of this work. Postdoc fellow Zhenhai Du, Ph.D. student Liangjun Hu, Zhuoning Zou, and Meishuo Liu from the School of Life Sciences at Tsinghua University are the co-first authors of this work. Prof. Yunlong Xiang from Chongqing Medical University, Clair Harris from the University of Michigan Medical School, Postdoc fellow Xukun Lu, Fengling Chen, Guang Yu, and Kai Xu, Ph.D. student Zihan Li and Feng Kong from the School of Life Sciences at Tsinghua University, also made significant contributions to this work. This study also received help from the Animal Center at Tsinghua University. This work is supported by the funding provided by the National Natural Science Foundation of China, the Key Research and Development Program of the Ministry of Science and Technology, the THU-PKU Center for Life Sciences, United States National Institutes of Health, and China Postdoctoral Science Foundation. Assistant Prof. Haifeng Wang is supported by Tsinghua University Initiative Scientific Research Program and the Benyuan Charity Fund. Prof. Wei Xie is a recipient of HHMI International Research Scholar award and New Cornerstone Investigator Award.
Full text:https://www.nature.com/articles/s41588-024-01897-2
Editor: Li Han

